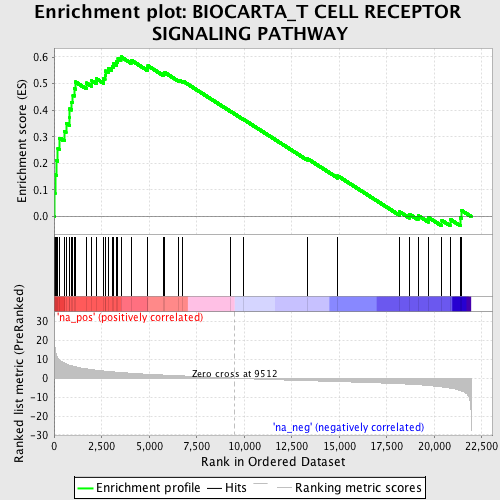
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_BT_ATM_minus_versus_ATM_plus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | BIOCARTA_T CELL RECEPTOR SIGNALING PATHWAY |
| Enrichment Score (ES) | 0.6005479 |
| Normalized Enrichment Score (NES) | 2.1243827 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0019299147 |
| FWER p-Value | 0.016 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | LCK | 20 | 19.234 | 0.0871 | Yes | ||
| 2 | CD3E | 51 | 15.588 | 0.1571 | Yes | ||
| 3 | NFKBIA | 116 | 12.486 | 0.2113 | Yes | ||
| 4 | CD247 | 200 | 10.587 | 0.2559 | Yes | ||
| 5 | CD4 | 297 | 9.537 | 0.2952 | Yes | ||
| 6 | PLCG1 | 539 | 7.963 | 0.3206 | Yes | ||
| 7 | VAV1 | 628 | 7.582 | 0.3513 | Yes | ||
| 8 | RAF1 | 819 | 6.824 | 0.3739 | Yes | ||
| 9 | NFATC1 | 826 | 6.800 | 0.4047 | Yes | ||
| 10 | RELA | 916 | 6.549 | 0.4306 | Yes | ||
| 11 | PPP3CB | 991 | 6.360 | 0.4563 | Yes | ||
| 12 | CD3D | 1059 | 6.186 | 0.4816 | Yes | ||
| 13 | ARHGAP4 | 1125 | 6.038 | 0.5062 | Yes | ||
| 14 | CAMK2B | 1700 | 4.893 | 0.5024 | Yes | ||
| 15 | MAPK3 | 1966 | 4.511 | 0.5110 | Yes | ||
| 16 | MAP3K1 | 2233 | 4.194 | 0.5180 | Yes | ||
| 17 | JUN | 2591 | 3.817 | 0.5192 | Yes | ||
| 18 | ARFGAP3 | 2680 | 3.722 | 0.5322 | Yes | ||
| 19 | SHC1 | 2716 | 3.694 | 0.5475 | Yes | ||
| 20 | PIK3CA | 2870 | 3.541 | 0.5567 | Yes | ||
| 21 | PTPRC | 3046 | 3.398 | 0.5643 | Yes | ||
| 22 | ZAP70 | 3141 | 3.317 | 0.5752 | Yes | ||
| 23 | ARHGAP1 | 3297 | 3.177 | 0.5826 | Yes | ||
| 24 | SOS1 | 3360 | 3.127 | 0.5941 | Yes | ||
| 25 | ARFGAP1 | 3521 | 3.003 | 0.6005 | Yes | ||
| 26 | CD3G | 4051 | 2.614 | 0.5884 | No | ||
| 27 | ELK1 | 4900 | 2.097 | 0.5592 | No | ||
| 28 | RALBP1 | 4938 | 2.076 | 0.5671 | No | ||
| 29 | PIK3R1 | 5770 | 1.654 | 0.5367 | No | ||
| 30 | MAP2K4 | 5823 | 1.626 | 0.5417 | No | ||
| 31 | FYN | 6571 | 1.275 | 0.5135 | No | ||
| 32 | FOS | 6777 | 1.183 | 0.5095 | No | ||
| 33 | LAT | 9292 | 0.110 | 0.3953 | No | ||
| 34 | NCF2 | 9952 | -0.199 | 0.3661 | No | ||
| 35 | MAP2K1 | 13318 | -1.307 | 0.2184 | No | ||
| 36 | ARHGAP6 | 14933 | -1.779 | 0.1529 | No | ||
| 37 | HRAS | 18182 | -2.929 | 0.0180 | No | ||
| 38 | PPP3CA | 18699 | -3.178 | 0.0090 | No | ||
| 39 | MAPK8 | 19156 | -3.453 | 0.0040 | No | ||
| 40 | ARHGAP5 | 19695 | -3.851 | -0.0030 | No | ||
| 41 | RAC1 | 20404 | -4.590 | -0.0143 | No | ||
| 42 | GRB2 | 20842 | -5.251 | -0.0102 | No | ||
| 43 | PRKCA | 21377 | -6.512 | -0.0048 | No | ||
| 44 | PPP3CC | 21439 | -6.721 | 0.0232 | No |